Today I am at the the symposium “Protein Chemistry ‐ Applications to Combat Diseases”, which takes place in Copenhagen a few minutes walk from where I work.
This morning started with a keynote lecture by Nobel Laurate Avram Hershko on regulation of the cell division cycle by ubiquitin‐mediated protein degradation. This post is just a very quick write-up and a few photos made during and immediately after his presentation.

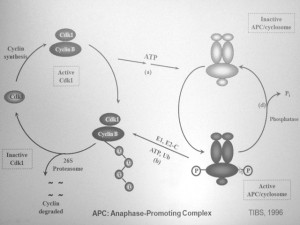
Most of the early work on ubiquitylation was done on model proteins, most of which were extracellular. Interestingly, what spurred Avram Hershko on to study ubiquitylation of physiologically relevant proteins was the early work on cyclin degradation for which Tim Hunt received the Nobel Prize. Tim Hunt speculated speculated that there was a cyclin protease that would break down cyclins. However, Avram Hershko showed in 1991 that cyclins are in fact not degraded by a specific protease, but are rather targeted for proteasomal degradation by a specific ubiquitin ligase. Showed this in JBC papers in 1991 and 1994. One year later his group identified this ubiquitin ligase to be what is now known as the Anaphase Promoting Complex (APC) / Cyclosome (APC/C).
In addition to being crucial for degradation of cyclins, APC/C is also required for entry into anaphase of the cell cycle (hence the name Anaphase Promoting Complex). This because it is responsible for targeting the securin protein for degradation, which in turns releases separase activity to degrade the cohesin rings that hold together sister chromatids.
Having worked on other cell-cycle proteins for many years, Avram Hershko has in recent years returned his interest to APC/C, more specifically to understand how the inhibition of APC/C is released, which in turn leads to the whole series of events described above.